Piecing together the Parkinson’s puzzle
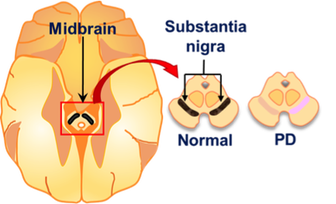
The movement disorder in Parkinson's disease (PD) is caused by a loss of dopamine neurons from the substantia nigra. PD is a complex multifactorial disease with roots in genetics, aging and the environment. Work in the Martin laboratory focuses on three main areas of investigation: (i) Genetic mutations that result in PD and provide important clues into the molecular basis of disease development (ii) genetic susceptibility to environmental toxins linked to PD and (iii) identifying mechanisms of biological aging that impact dopamine neuron health and vulnerability to loss in PD pathogenesis. For each area, we use a combination of Drosophila and rodent animal models and a wealth of biochemical, advanced imaging, -omics and behavioral approaches.
The movement disorder in Parkinson's disease (PD) is caused by a loss of dopamine neurons from the substantia nigra. PD is a complex multifactorial disease with roots in genetics, aging and the environment. Work in the Martin laboratory focuses on three main areas of investigation: (i) Genetic mutations that result in PD and provide important clues into the molecular basis of disease development (ii) genetic susceptibility to environmental toxins linked to PD and (iii) identifying mechanisms of biological aging that impact dopamine neuron health and vulnerability to loss in PD pathogenesis. For each area, we use a combination of Drosophila and rodent animal models and a wealth of biochemical, advanced imaging, -omics and behavioral approaches.
|
LRRK2 in Parkinson's disease
Mutations in LRRK2 (leucine-rich repeat kinase 2) are commonly found in both familial and sporadic PD. Preventing LRRK2-induced neurodegeneration will require a detailed understanding of the key mechanisms driving neuronal dysfunction and death. LRRK2 has been implicated in numerous biological processes, but defining mechanisms that drive age-related neuronal death has been elusive. We performed a comprehensive screen to identify genes that influence age-dependent loss of dopamine neurons in LRRK2 G2019S-expressing Drosophila. This approach yielded a number of genes that regulate the morphology of neurites (axon and dendrites). We are working with Drosophila and mammalian disease models to delineate the nature of mutant LRRK2-induced neurite defects in vivo, their underlying cause, and their relationship to dopamine neuron death. |
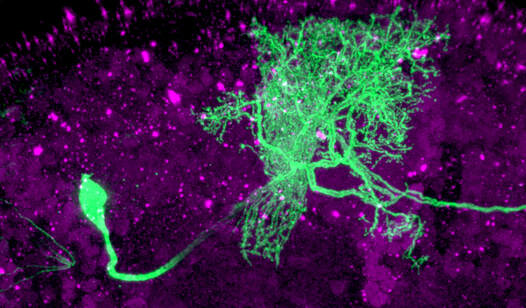
Single GFP-labeled dopamine neuron showing axon terminal arborization. Maximum projection image through the posterior Drosophila brain following confocal microscopy.
|
Role of aging in neurodegeneration
The biggest risk factor for developing PD is age, suggesting that age-related changes in the brain predispose to loss of dopamine (DA) neuron health and viability. Cell stressors such as oxidative stress and metabolic dysfunction increase with age, and DA neurons are particularly vulnerable to this stress. A growing body of evidence indicates that oxidative stress-responsive signaling may promote aging, although the molecular mediators are not well understood. We are actively investigating how oxidative stress-responsive signaling mediators important for lifespan regulation intersect with neuronal health across age, particularly age-related maintenance of dopamine neuron viability.
The biggest risk factor for developing PD is age, suggesting that age-related changes in the brain predispose to loss of dopamine (DA) neuron health and viability. Cell stressors such as oxidative stress and metabolic dysfunction increase with age, and DA neurons are particularly vulnerable to this stress. A growing body of evidence indicates that oxidative stress-responsive signaling may promote aging, although the molecular mediators are not well understood. We are actively investigating how oxidative stress-responsive signaling mediators important for lifespan regulation intersect with neuronal health across age, particularly age-related maintenance of dopamine neuron viability.
Genetic susceptibility to neurotoxins in PD
Epidemiological studies link pesticide exposure to PD risk, yet human studies alone have not conclusively linked PD to any single pesticide. Controlled animal models play a vital role in determining whether pesticides can cause PD-related neurodegeneration and can be leveraged to gain mechanistic insight into disease etiology. We are using Drosophila to identify genes underlying susceptibility to neurotoxins such as pesticides that have been linked to PD. Genes identified through this approach will then be pursued in rodent models of neurodegeneration in order to assess their potential contribution to disease development.
.
Epidemiological studies link pesticide exposure to PD risk, yet human studies alone have not conclusively linked PD to any single pesticide. Controlled animal models play a vital role in determining whether pesticides can cause PD-related neurodegeneration and can be leveraged to gain mechanistic insight into disease etiology. We are using Drosophila to identify genes underlying susceptibility to neurotoxins such as pesticides that have been linked to PD. Genes identified through this approach will then be pursued in rodent models of neurodegeneration in order to assess their potential contribution to disease development.
.